Plant genome structure and evolution, especially the nature of rearrangements
and the contributions of transposable elements
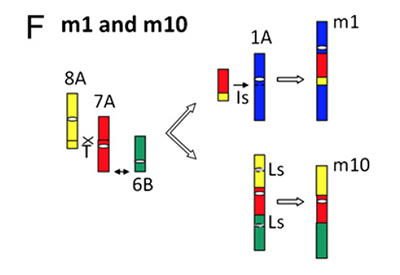
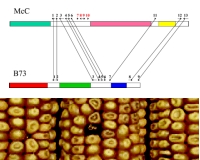
Molecular mapping using DNA probes, and now comprehensive DNA sequencing projects, have shown that gene content and order are significantly conserved between species, especially in closely related species. However, complex plant genomes like those in the angiosperms are relatively unstable, rearranging gene content and order at rates as much as 100-fold faster than mammalian genomes, for instance. We have begun to define the nature of these rearrangements and their mechanisms. Our preliminary data indicate that transposable elements are particularly clear sensors of these changes, many of which are caused by ectopic homologous recombination and illegitimate recombination. Ongoing and future studies will precisely determine the mechanisms of illegitimate recombination, will define the differences in the frequencies and types of these events in different plant lineages, and will measure how these processes contribute to the C value paradox. Bioinformatics and high throughput genomics are the key tools in these studies.
The relationship between genome structure/evolution and gene function
We are only beginning to understand how genome context (that is, the nature of the DNA/chromatin that surrounds genes) can influence gene function and gene evolution. The grasses that we study are particularly well-suited to such endeavors because genome structure changes so rapidly. We are using the full set of genomics tools, including bioinformatics, full genome sequencing, expression analyses and reverse genetics, to try to determine the relationship between genomic context and gene expression. In addition, we are using these same tools to answer one of the great questions in biology. Namely, what are the specific genetic changes that differentiate species? In order to pursue these studies from a strong foundation, we need to have full genome sequence information for many species of plants. Current approaches to genome sequencing are too expensive to permit analysis of more than a few species, so we have developed gene-enrichment technologies that will allow low cost and very quick sequencing of any complex genome (plant, animal, fungal or protist). These technologies are now being tested in a project to sequence and orient the gene space within the maize nuclear genome.
Genetic diversity and its use in under-utilized crops of the developing world

Modern crop improvement has led to the severe narrowing of germplasm diversity, both within species of crops and in the sets of crops that are now extensively utilized. Highly productive and stress-adapted cereals like tef (Eragrostis tef), finger millet (Eleusine indica) and foxtail millet (Setaria italica) are often replaced by less-adapted species like maize and wheat because of marketing forces. These "minor cereals" tend to be vital mainly to subsistence farmers in Africa and other parts of the developing world, so they have received little research attention. We are working on three subsistence cereals: tef, finger millet, and highland rice. Because they are so under-studied, the potential for significant improvement is much higher than for intensively studied/improved species like maize or lowland rice. Our efforts are mainly associated with studies of genetic diversity, and particularly any diversity that associates with useful agronomic properties like abiotic or biotic stress tolerance. A major component of this project involves the transfer of molecular technologies into developing world laboratories.
The rapid evolution of complex disease resistance loci in plants

Plants have many disease resistance genes that initiate a signal transduction cascade of antimicrobial factors after the pathogen signal is recognized. Many of these resistance genes are highly complex and highly unstable, often in ways that are comparable to events seen at the immunoglobin and histocompatibility genes of chordates. We are studying one of these genes, Rp1 of maize, in significant detail. We have sequenced the entire complex of more than 650 kb. We are now characterizing gene expression, gene function, and the mechanisms of hyper-evolution in this cluster. In addition, we are studying and isolating other disease resistance genes from maize and sorghum. These new resistance genes will provide an interesting comparison to the Rp1 complex and may also prove useful for crop improvement.
Models, mechanism and metagenomics for lignocellulosic ethanol production

Plants have evolved an efficient process, photosynthesis, for conversion of sunlight into usable energy. Most of the energy available for human use (coal, petroleum, and firewood) is derived from either ancient or current photosynthesis. Although the use of most biofuels is currently carbon negative or carbon neutral (that is, as much or more carbon is released into the atmosphere than is captured in the fuel), lignocellulosic energy conversion of perennial grasses like switchgrass has at least a seven-fold benefit in carbon capture, mostly by the roots and rhizosphere of the unharvested portions of the plants. We are involved in several consortia of projects that will attempt to improve the efficiency of lignocellulosic ethanol production so that it will be less expensive than current petroleum-based projects. The approach taken has three components. The first is development of foxtail millet (Setaria italica), an important grain of Northern China and Mongolia, as a model for switchgrass and other perennial grasses. We are collaborating with the Joint Genome Institute (DOE) in the sequencing and annotation of this genome. We are also working with other groups to improve the transformation, genetic maps and other components of the genetic/genomic toolkit for this species. Second, we are cloning and characterization genes from switchgrass and foxtail millet that are known to be involved in both the quality and quantity of biomass produced by these species. Third, and finally, we are employing metagenomic analysis of the microbial components of the switchgrass rhizosphere to find the microbes that will help sustain and promote high biomass quality and yields in this perennial crop.